Byssocorticium pulchrum (S. Lundell) M.P. Christiansen
Beautiful Blue Crust
Basidiomycota: Agaricomycotina: Agaricomycetes: Atheliales: Byssocorticiaceae:![]() Byssocorticium
Byssocorticium![]()
![]()
Introduction
Like its cousin Byssocorticium atrovirens, B. pulchrum is a beautiful blue crust found under wood and leaves in the forest. Indeed, its Latin name "pulchrum" translates to "beauty". To differentiate the two you will need to look at their spores under the microscope. B. pulchrum has slightly larger spores (5–6 µm in diameter) compared to B. atrovirens (3–4 µm). This small morphological difference corresponds well to molecular phylogenetic groupings.
Byssocorticium in general is poorly represented in sequence databases. Just three other vouchered specimens identified as B. pulchrum have been sequenced and all of them are from northern Europe. The ITS region is only about 93-95% identical between this New Jersey specimen and two Swedish specimens. Examining the ITS tree below, B. pulchrum from the Americas may constitute a distinct species from European and Asian B. pulchrum. This parallels B. atrovirens, which also shows high ITS divergence across geographic regions. However, in both cases the species complexes appear to have a consistent morphology. At the end of the day splitting these taxa or keeping them lumped might just be an arbitraty decision, although more sequences could help clear things up.
Description
Ecology: Mycorrhizal, growing on the underside of wood debris.
Basidioma: Effused, byssoid to athelioid, with an even to weakly merulioid hymenophore and an indeterminate, arachnoid margin; bright, light chalky blue when fresh, fading slightly when dried.
Chemical reactions: NA
Spore print: NA
Hyphal system: Monomitic, simple septa and clamps both present, greenish in Melzer's reagent; (3.1) 3.3–3.8 (3.9) µm wide (n = 10).
Basidia: Terminal, clavate, with four sterigmata, occassionally with guttules and sometimes with a basal clamp; length (15.4) 17.6–23.3 (24.3) µm, width (4.6) 5.1–6.5 (6.7) µm, x̄ = 20.5 ✕ 5.8 µm (n = 10).
Basidiospores: Globose to subglobose, smooth, hyaline, apiculate; length (4.9) 4.9–5.6 (6) µm, width (4.4) 4.7–5.3 (5.9) µm, x̄ = 5.3 ✕ 5.0 µm, Q (1) 1–1.1 (1.2), x̄ = 1.1 (n = 30).
Sterile structures: Absent.
Sequences: ITS rDNA (ON364097), LSU rDNA (ON369550).
Notes: All measurements taken in 5% KOH stained with phloxine.
Specimens Analyzed
ACD0471, iNat98605664; 17 October 2021; Estelle Manor Park, Atlantic Co., NJ, USA, 39.4145 -74.7321; leg. Alden C. Dirks, det. Alden C. Dirks, ref. Jülich & Stalpers (1980) and Bernicchia & Gorjón (2010); University of Michigan Fungarium ——.
References
Bernicchia, A. & Gorjón, S.P. (2010). Fungi Europaei, Volume 12: Corticiaceae s.l. Massimo Candusso, Italia.
Jülich, W. & Stalpers, J. A. (1980). The Resupinate Non-poroid Aphyllophorales of the Temperate Northern Hemisphere. North-Holland Publishing Company, Amsterdam, Oxford, New York.
Links

The beautiful blue crust forms gorgeously colored basidiomata with a slightly merulioid texture when fresh.

The hymenophore is loosely attached to the substrate as a delicate, membranaceous sheath that can be removed like Athelia, although it tears easily.
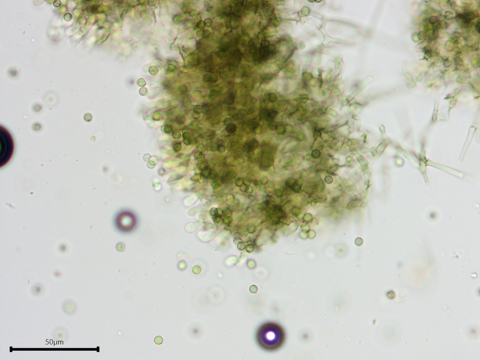
The hyphae and spores are greenish in Melzer's reagent.
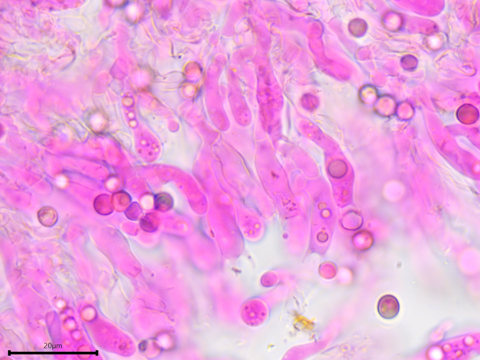
The basidia are somewhat clavate and sometimes contain guttules.
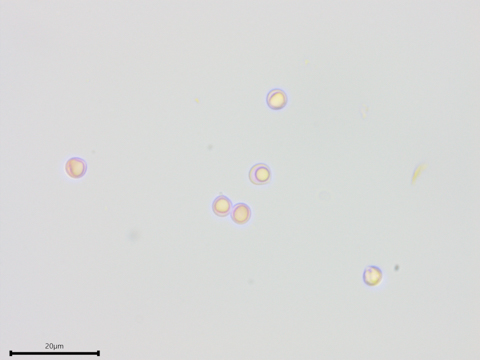
Distinctly apiculate, subglobose spores with a single large guttule.
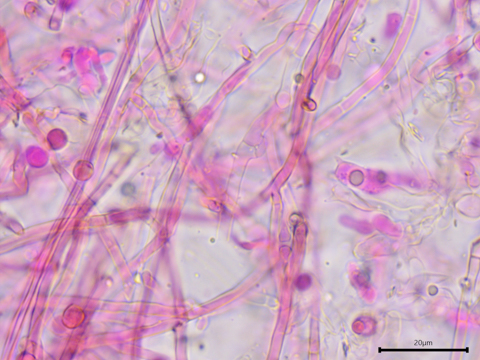
Infrequently clamped monomitic hyphal system.
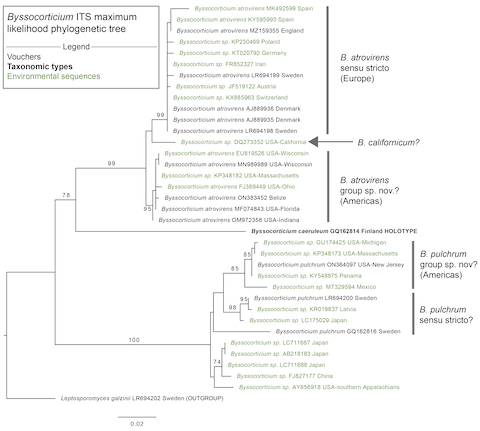
Byssocorticium ITS maximum likelihood phylogenetic tree. The sequences were aligned in SeaView with MUSCLE, trimmed with TrimAl, and analyzed with RAxML using 1000 bootsrap replicates and the GTR+G model. Sequences are annotated with their GenBank accession number and location of origin. Bootstrap values ≥ 70% are considered significant. Leptosporomyces galzinii serves as the outgroup.
